First ever Kaggle competition - Pneumonia Detection
Github here: https://github.com/varunvprabhu/pneumonia_detection
I'd never participated in a Kaggle competition before. So I decided to join one, namely, the RSNA Pneumonia Detection challenge.
I'd never participated in a Kaggle competition before. So I decided to join one, namely, the RSNA Pneumonia Detection challenge.
Briefly, the competition was about developing an AI algorithm that would assist radiologists in pneumonia detection.
The dataset training and test images were provided by the competition organizers through Kaggle. Approximately 28000 training images and 1000 test images were provided. Some of the 28000 images had bounding boxes of the locations of pneumonia detections in chest x-rays.(Specifically 8964 images)
The 1000 images were un-labeled and one had to post the results of the algorithm after the images were run through that. A score is calculated using the mean taken over the individual average precision of each image in the test dataset.
There were 2 ways to go about this. One was through object detection and the other was through segmentation. For object detection, YOLO immediately came to mind. However, I wasn't sure if it was a best fit for this competition. I thought that the pneumonia detections within the x-rays didn't have enough features so that it could be detected accurately enough by the algorithm. From some of the public kernels though, I could see that people had tried YOLO and had obtained decent results. This was from transfer learning models.
I chose the second method, namely segmentation. I'd previously used the hourglass model to segment body parts for pose detection. I decided to use the same for this competition.
The process I used was similar to the previous project. I created masks for each image where pneumonia was detected. In cases where it wasn't detected, I created an all black image to indicate zero detections. Then I ran the original images through the network and used a square loss function against the masks to train the network to detect pneumonia instances.
However this post gave some interesting insights into the dataset. Basically, there were 2 kinds of negative labels. One was a set of normal images that had no lung diseases whatsoever. The second set consisted of lung xrays that had diseases but were not pneumonia specifically. I think this made the detections a bit more difficult since some of the abnormal xrays seemed to have features similar to pneumonia but weren't that at all. Moreover there were no bounding boxes provided for these with explicit labels for the different lung diseases. The only info provided was that they were abnormal. Initially I didn't change anything in the masks to take this info into account.
The post processing that I used for the network output images was: thresholding followed by connected components. Another discriminator that I used was the area size of the boxes given as an output by the connected components algorithm. I used those two parameters to get a local maxima with the scores.
Thresholding with a higher pixel value eliminated some smaller sized false positives but also reduced precision with respect to area sizes. A much smaller threshold discovered too many areas. Reducing the area size taken into consideration would improve smaller detections but would also bring in false positives.
When I ran the algorithm the first time, I only used the masks from the bounding boxes provided from the pneumonia images and got a score of 0.124 which squarely put me in the slightly above average scores. I used a thresholding of 40 and an area size of 64.
The second time around, I decided to use an additional generic mask for the abnormal images. I figured that would help eliminate some false positives from the test images. Since bounding boxes weren't provided for these images, I used a general bounding box size based on the heatmap value of detections and made a mask out of that.
Using a threshold of 50 with area size of 256, I managed to improve the score to 0.134. Again, a slightly above average score.
I also tried having a rolling threshold where threshold values were iterated through an array and if the connected components managed to find more than 2 areas, then those values were recorded. However this did not improve the score.
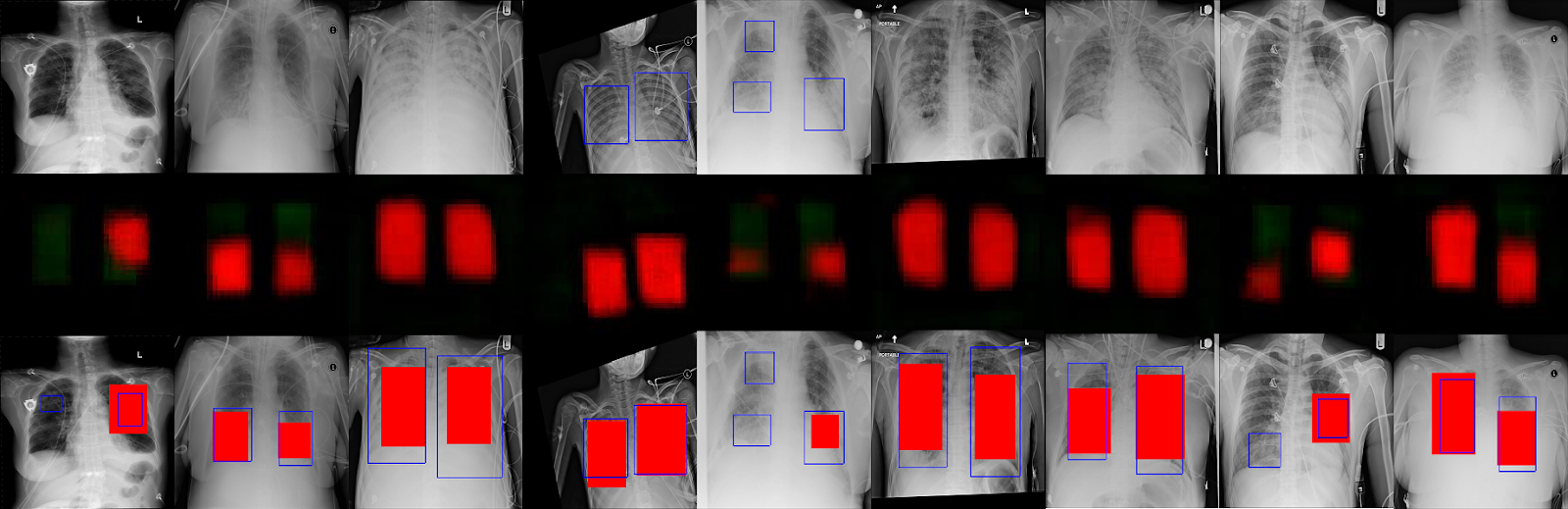
Some of the results are below. Ill update this post with the final standings or algorithm updates once the competition is over.
First row has the original chest xray images. Second row has the predictions from the neural network. Third row has the predicted boxes in red along with the actual bounding boxes in blue. In some cases, the boxes are a good match. In other cases, there are no detections, especially with the smaller boxes.
Update 2018-10-17: I tried an ensemble method of generating results. What I did was:
1) I trained a new model where I performed image augmentations on the training set during training. I rotated the images and added a slight gaussian blur (separately). The model outputs at a threshold value of 60 and area size of 256 for connected components gave me a max score of 0.130. Let's called this Model B.
2) Then I combined the results of this model with that of the model that gave me a score of 0.134. Let's call the model that gave this score of 0.134 Model A.
3) The combination worked as follows: I generated csv files from both models for the test set at the best threshold and area values. If model B predicted more boxes than model A for a specific image, the data was copied over to model A. Also if model B predicted data for an image but model A didn't, the data was copied over to Model A. Any other model A data was untouched.
4) This algorithm above gave me a max score of 0.154!
5) I trained the model B for some more epochs and again chose an output with threshold 60 and area 256. The individual score was 0.126. Combined with model A, I got a new high score of 0.157! This was my best score so far and in the standings as of this writing, I was at position 300 out of 1300.
Update 2018-11-04: Competition is over and my final position is 236 out of 346 teams that submitted models for stage 2. The winning submission used an ensemble of models. They used a detection model instead of segmentation model.
Update 2018-11-06: one of the top models used a simpler ensemble model that used retinanet. It appears that the object detection models performed the best. Guess I'll need to read about retinanet and implement it sometime in the future. Here's the link to that submission:
https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/discussion/70632
I also tried having a rolling threshold where threshold values were iterated through an array and if the connected components managed to find more than 2 areas, then those values were recorded. However this did not improve the score.
Some of the results are below. Ill update this post with the final standings or algorithm updates once the competition is over.
First row has the original chest xray images. Second row has the predictions from the neural network. Third row has the predicted boxes in red along with the actual bounding boxes in blue. In some cases, the boxes are a good match. In other cases, there are no detections, especially with the smaller boxes.
Update 2018-10-17: I tried an ensemble method of generating results. What I did was:
1) I trained a new model where I performed image augmentations on the training set during training. I rotated the images and added a slight gaussian blur (separately). The model outputs at a threshold value of 60 and area size of 256 for connected components gave me a max score of 0.130. Let's called this Model B.
2) Then I combined the results of this model with that of the model that gave me a score of 0.134. Let's call the model that gave this score of 0.134 Model A.
3) The combination worked as follows: I generated csv files from both models for the test set at the best threshold and area values. If model B predicted more boxes than model A for a specific image, the data was copied over to model A. Also if model B predicted data for an image but model A didn't, the data was copied over to Model A. Any other model A data was untouched.
4) This algorithm above gave me a max score of 0.154!
5) I trained the model B for some more epochs and again chose an output with threshold 60 and area 256. The individual score was 0.126. Combined with model A, I got a new high score of 0.157! This was my best score so far and in the standings as of this writing, I was at position 300 out of 1300.
Update 2018-11-04: Competition is over and my final position is 236 out of 346 teams that submitted models for stage 2. The winning submission used an ensemble of models. They used a detection model instead of segmentation model.
Update 2018-11-06: one of the top models used a simpler ensemble model that used retinanet. It appears that the object detection models performed the best. Guess I'll need to read about retinanet and implement it sometime in the future. Here's the link to that submission:
https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/discussion/70632

Comments
Post a Comment